Minimal Neighborhood Redundancy Maximal Relevance: Application to the Diagnosis of Alzheimer's Disease
Abstract
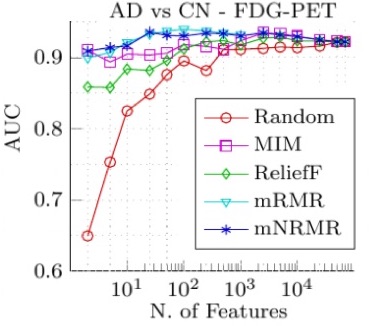
Existing feature selection methods are able to choose discriminative features with low redundancy but are computationally too expensive for neuroimaging applications. This occurs because they analyze every brain voxel while trying to reduce the redundancy between the selected features. We propose a significantly faster method that focuses on the main source of redundancy which is neighboring voxels and compare this new approach with four other well-known feature selection methods, evaluating them extensively on three datasets. We start by using an artificial dataset to study the robustness of our approach to noisy features, erroneous labels and small number of samples, which are problems that are often encountered when building a CAD system that takes brain images as its input. Then, we analyze the computational complexity of our method and study its usefulness for the diagnosis of Alzheimer’s disease and Mild Cognitive Impairment using FDG-PET images and tissue probability maps of Gray-Matter extracted from MR images. Experimental results on synthetic and real data clearly validate our approach as a very efficient algorithm for the selection of non-redundant features applicable to a variety of neuroimaging techniques. In fact, the major computational gains come at no cost in either performance or robustness.
Published at: Neurocomputing, Vol. 155, pp. 295-308, May, 2015.
Bibtex
@article{morgado2015minimal,
title={Minimal neighborhood redundancy maximal relevance: Application to the diagnosis of Alzheimer׳ s disease},
author={Morgado, Pedro M and Silveira, Margarida and Alzheimer׳ s Disease Neuroimaging Initiative and others},
journal={Neurocomputing},
volume={155},
pages={295--308},
year={2015},
publisher={Elsevier}
}